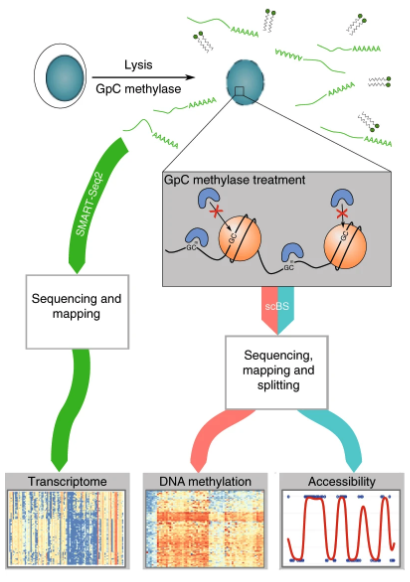
The scNMT-seq uses the similar principle as scNOMe-seq, where a purified GpC methylase (M.CviPI) was applied on permeabilised cells. The majority of the DNA is protected by nucleosomes and inaccessible to M.CviPI. Only GpC within accessible regions can be methylated by M.CviPI. Then, mRNA is dragged out by oligo-dT magnetic beads, and library preparation was done using the SMART-seq2 method, click here to see a step-by-step protocol of SMART-seq2. After bisulfite conversion of the gDNA, the scBS-seq method was used to prepare the library to look at methylation (CpG) and chromatin accessibility (GpC), click here to see a step-by-step protocol of scBS-seq.