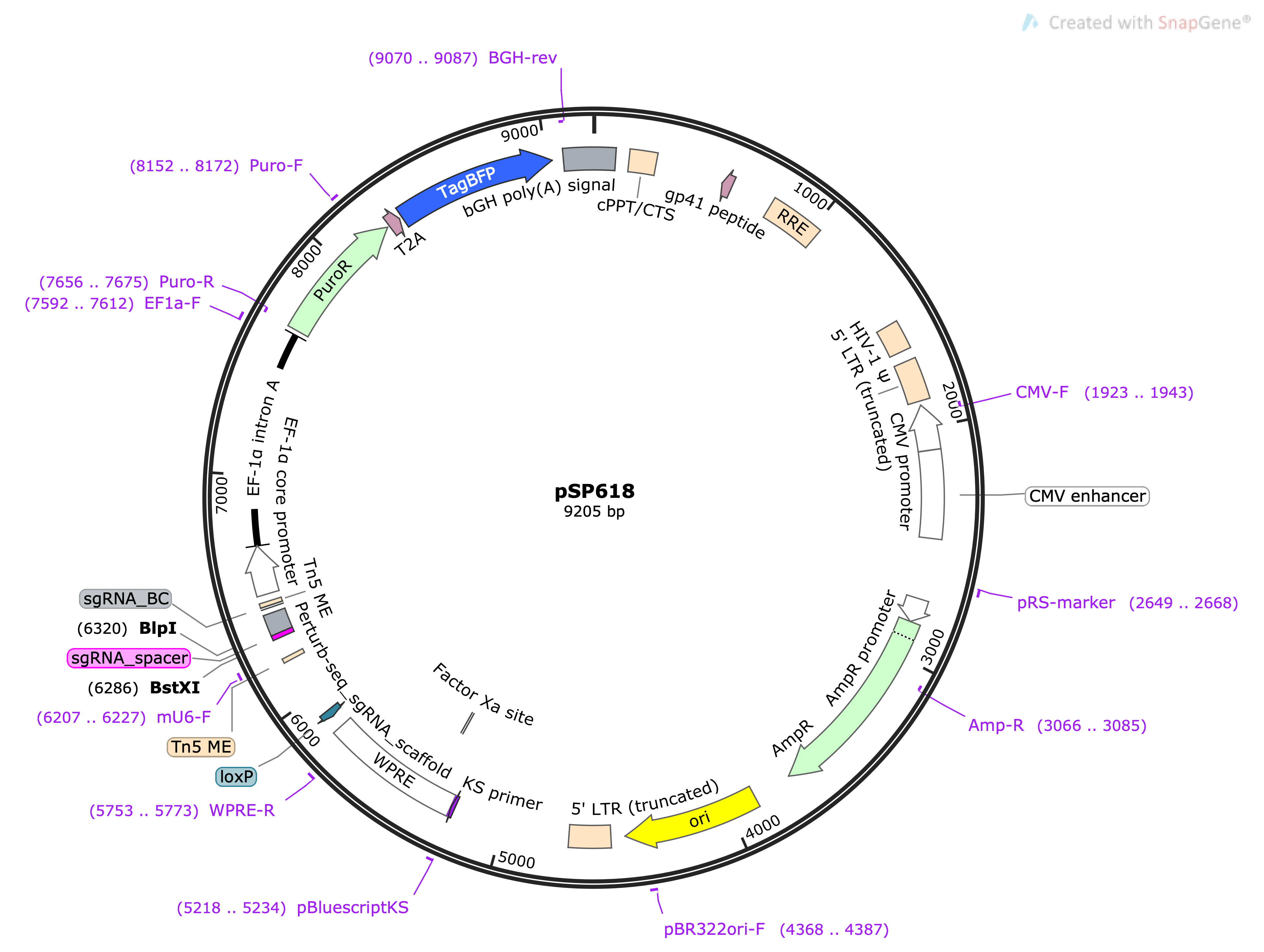
|--BstXI---| |BlpI | 5'- ctag CCACCTTGTTGG [sgRNA-Spacer] GTTTAAGAGCTAAGC ctag gatcGGTGGAACAACC [sgRNA-Spacer] CAAATTCTCGATTCG gatc -5'
|BlpI | |--BstXI---| 5'-[other part of the vector backbone]- TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGAAATCCAAGCCTATCATGTAAAATGTAGC -[other part of the vector backbone]-3' 3'-[other part of the vector backbone]-AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG TCTTTAGGTTCGGATAGTACATTTTACATCG -[other part of the vector backbone]-5's5 ME Perturb-seq sgRNA scaffold <------ mU6-F ------- ME s7
oSP1735: 5'-
oSP2053: 5'-/Bio/
oMCB1672: 5'-
Beads-oligo: |--5'-
SI-PCR Primer B (PN-2000128): 5'-
i7 Sample Index Plate N, Set A (PN-3000262): 5'-
P7-TruSeqR2: 5'-
Nextera left primer entry point (s5): 5'-
Nextera right primer entry point (s7): 5'-
Illumina Nextera Read 1 primer: 5'-
Illumina Nextera Read 2 primer: 5'-
ATAC sample index sequencing primer (index1): 5'-
sgRNA sample index sequencing primer (index1): 5'-
Cell barcode sequencing primer (index2): 5'-
Illumina P5 adapter: 5'-
Illumina P7 adapter: 5'-
Product 1 (s5 at both ends, not amplifiable due to semi-suppressiev PCR): 5'- TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA CTGCGACGGCTGCT -5' Product 2 (s7 at both ends, not amplifiable due to semi-suppressiev PCR): 5'-GTCTCGTGGGCTCGG AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5' Product 3 (different ends, amplifiable): 5'-TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5'
Only the relevant bits around the sgRNA is shown (may get tagged by Tn5 but the probability is low): 5'- ... TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGAAATCCAAGCCTATCATGTAAAATGTAGC ... -3' 3'- ...AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG TCTTTAGGTTCGGATAGTACATTTTACATCG ... -5'
|--5'- AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC --------------> 5'-TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXXXXXXXXXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXXXXXXXXXXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5'
|--5'- AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC --------------> 5'- ...TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGAAATCCAAGCCTATCATGTAAAATGTAGC ... -3' 3'- ...AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG TCTTTAGGTTCGGATAGTACATTTTACATCG ... -5' <----------GGTTCGGATAGTACATTTTACATCG -5'
ATAC: 5'- AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXXXXXXXXXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXXXXXXXXXXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5' sgRNA: 5'-AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGAAATCCAAGCCTATCATGTAAAATGTAGC -3'TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG TCTTTAGGTTCGGATAGTACATTTTACATCG -5'
ATAC: 5'- AATGATACGGCGACCACCGAGA ----------------> 5'-AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5' <----------------GGCTCGGGTGCTCTG [8-bp sample index]TAGAGCATACGGCAGAAGACGAAC -5' sgRNA: 5'-AATGATACGGCGACCACCGAGA ----------------> 5'-AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC AGAAATCCAAGCCTATCATGTAAAATGTAGC -3'TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG TCTTTAGGTTCGGATAGTACATTTTACATCG -5' <----------------GGCTCGGGTGCTCTG [8-bp sample index]TAGAGCATACGGCAGAAGACGAAC -5'
ATAC (ready to sequence): 5'- AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC [8-bp sample index]ATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG [8-bp sample index]TAGAGCATACGGCAGAAGACGAAC -5' sgRNA (the library is currently dominated by ATAC, we still need further PCR to specifically enrich the sgRNA library): 5'-AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC [8-bp sample index]ATCTCGTATGCCGTCTTCTGCTTG TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG [8-bp sample index]TAGAGCATACGGCAGAAGACGAAC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTATAGTCTCAAAACACACAATTACTTTACAGTTAGGGTGAGTTTCCTTTTGTGCTG TTTCTGTCTCTTATACACATCT CCGAGCCCACGAGAC [8-bp sample index]ATCTCGTATGCCGTCTTCTGCTTG TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATATCAGAGTTTTGTGTGTTAATGAAATGTCAATCCCACTCAAAGGAAAACACGAC AAAGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG [8-bp sample index]TAGAGCATACGGCAGAAGACGAAC -5' <---------GTTCCACC AAGAGGTTCCCTATGAATCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG /Bio/-5'
5'- AATGATACGGCGACCACCGAGA ----------------> 5'-AATGATACGGCGACCACCGAGATCTACAC [16-bp cell barcode] TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG [10-bp sgRNA barcode] CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC TTACTATGCCGCTGGTGGCTCTAGATGTG [16-bp cell barcode] AGCAGCCGTCGCAG TCTACACATATTCTCTGTC [10-bp sgRNA barcode] GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG -5' <------------GTGTGCAGACTTGAGGTCAGTG [8-bp i7]TAGAGCATACGGCAGAAGACGAAC -5'
ATAC: 5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNNNNNNNNNN TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'Illumina P5 16-bp cell s5 ME gDNAME s7 8-bpIllumina P7 barcode sample index sgRNA: 5'-AATGATACGGCGACCACCGAGATCTACAC NNNNNNNNNNNNNNNN TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG NNNNNNNNNN CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC NNNNNNNNNN GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'Illumina P5 16-bp cell s5 ME 10-bp Perturb-seq sgRNA scaffold TruSeq Read 2 8-bpIllumina P7 barcode sgRNA barcode sample index
5'- TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG -------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
5'- CTGTCTCTTATACACATCT CCGAGCCCACGAGAC -------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNNNNNNNNNN TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG -3' <---------------AGCAGCCGTCGCAG TCTACACATATTCTCTGTC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNNNNNNNNNN TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT CCGAGCCCACGAGAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG -3' <------GACAGAGAATATGTGTAGA GGCTCGGGTGCTCTG -5'
5'- GCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC ------------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC NNNNNNNNNN GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
5'- AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNNNNNNNNNN AGCAGCCGTCGCAG TCTACACATATTCTCTGTC NNNNNNNNNN GCTCCGACTCACATCTAAGCTCGTTTTTCGTGGCTGAGCCACGGTGAAAAAGTTCAACTATTGCCTGATCGGAATAAATTTGAACGATACGACAAAGGTCGAATCGAGAATTTG [sgRNA-Spacer] GGTTGTTCCACC AAGAGGTTCCCTATGAATCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNNNNNNNNNN TCGTCGGCAGCGTC AGATGTGTATAAGAGACAG NNNNNNNNNN CGAGGCTGAGTGTAGATTCGAGCAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCTTAGCTCTTAAAC [sgRNA-Spacer] CCAACAAGGTGG TTCTCCAAGGGATACTTAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG <---------------AGCAGCCGTCGCAG TCTACACATATTCTCTGTC -5'