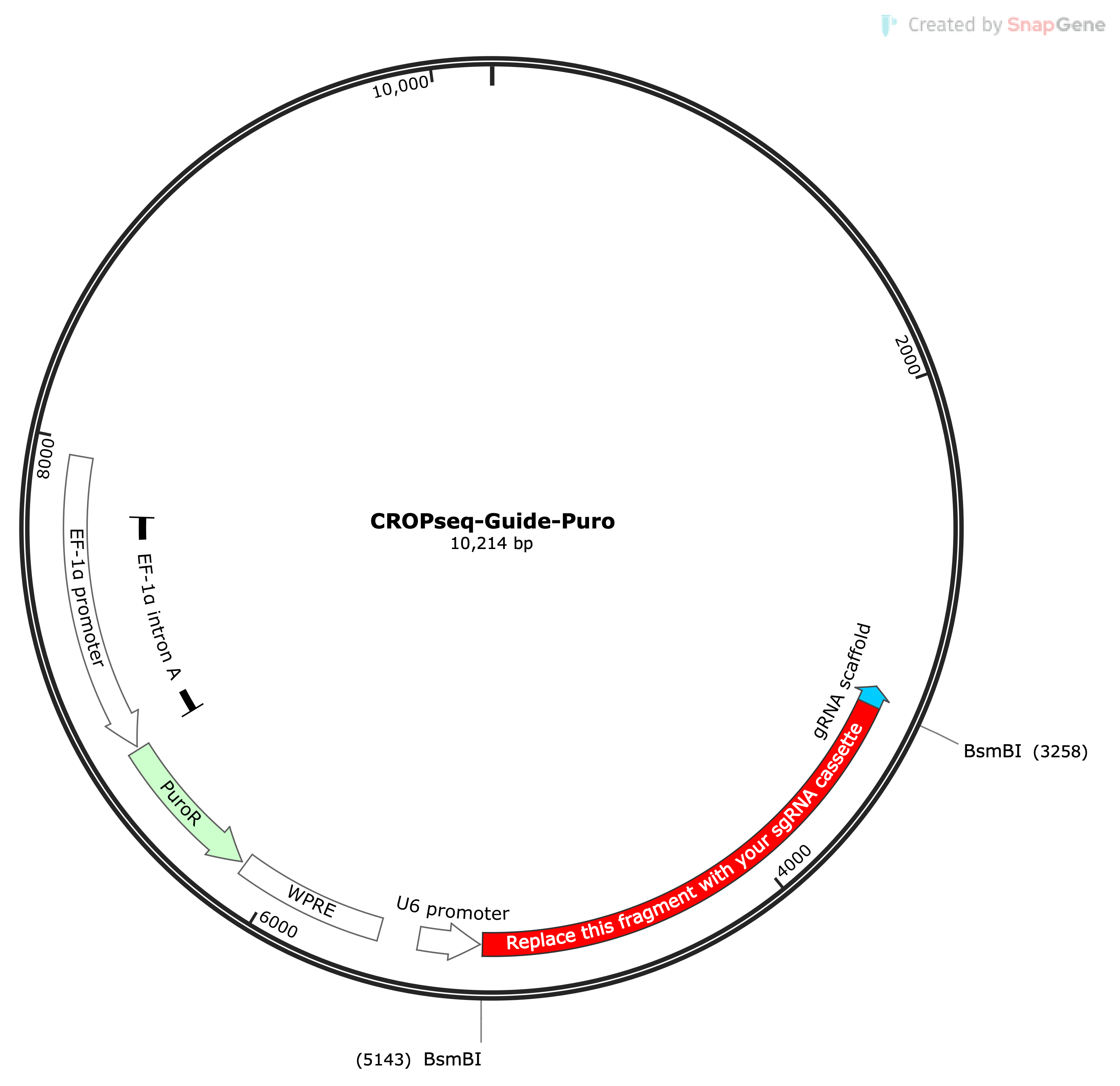
CRISPR-sciATAC is a multi-omic method to perform pooled sgRNA screen using single-cell ATAC-seq as readout. The data give you the chromatin accessibility profile of each single cell and the sgRNA information from the same cell. You can think of this as the the pooled version of Purterb-ATAC. It uses the CROP-seq vector backbone for the sgRNA, meaning it profiles the sgRNA via the transcripts. The authors also identified a new ViPar transposase with mutations P50K and M53Q, which they called TnY, and it gives higher yield during protein purification and has similar tagmentation activity compared to the commonly used Tn5. Then they use TnY with the combinatorial indexing strategy to achieve high cell throughput.
The workflow drawn in the page is based on their Supplementary Figure 1. The full barcoded oligo sequences can be found from their Supplementary Table 2.
5'- CACCG [sgRNA-Spacer] C [sgRNA-Spacer] CAAA -5'
5'-[PuroR]-[WPRE]- ...UGCAUAUACGAUACAAGGCUGUUAG...UUGUGGAAAGGAC GAAACACCG [sgRNA-Spacer] GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUU -[Poly-A] -3'|-------------- U6 promoter ---------------| CROP-seq sgRNA scaffold
pMENT (ME): 5'-/Phos/
T5 tagment sciATAC: 5'-
Barcoded T7 tagment ATAC: 5'-
Barcoded RTi sgRNA: 5'-
Barcoded P5i ATAC PCR1: 5'-
Barcoded P7i ATAC PCR1: 5'-
Barcoded P5i sgRNA PCR1: 5'-
Staggered P5 sgRNA PCR2: 5'-
Barcoded P7i sgRNA PCR2: 5'-
Read 1 ATACseq: 5'-
Index 1 ATACseq: 5'-
Read 2 ATACseq: 5'-
TruSeq Read 1: 5'-
Illumina P5 adapter: 5'-
Illumina P7 adapter: 5'-
Product 1 (T5 at both ends, not amplifiable due to semi-suppressiev PCR): 5'- TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA CTCCGCCTCTGCCAC GCAACCAACGCACCTCTGCGACGGCTGCT -5' Product 2 (T7 at both ends, not amplifiable due to semi-suppressiev PCR): 5'-GTCTCGTGGGCTCGGCTGTCCCTGTCC [8-bp barcode1] GCGATCGAGGACGGC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG [8-bp barcode1] CCTGTCCCTGTCGGCTCGGGTGCTCTG -5' Product 3 (different ends, amplifiable): 5'-TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG [8-bp barcode1] CCTGTCCCTGTCGGCTCGGGTGCTCTG -5'
Anneal RT primer and reverse transcription: 5'- CCGGGAGCTGCATGT [8-bp barcode1] TTTTTCAAGTTGATAA ----------------------> 3'- [Poly-A]-UUUUUCGUGGCUGAGCCACGGUGAAAAAGUUCAACUAUUGCCUGAUCGGAAUAAAAUUGAACGAUAAAGAUCGAGAUUUUG [sgRNA-Spacer] GCCACAAAG CAGGAAAGGUGUU...GAUUGUCGGAACAUAGCAUAUACGU... -[WPRE]-[PuroR] -5' This the first strand cDNA: 5'-CCGGGAGCTGCATGT [8-bp barcode1] TTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC [sgRNA-Spacer] CGGTGTTTC GTCCTTTCCACAA...CTAACAGCCTTGTATCGTATATGCA... -[WPRE]-[PuroR] -3'
5'- AATGATACGGCGACCACCGAGATCTACAC [8-bp barcode2] TCGTCGGCAGCGTC -----------> 5'-TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXXXXXXXXXXX...XXXCTGTCTCTTATACACATCT TCTACACATATTCTCTGTC XXX...XXXXXXXXXXXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG [8-bp barcode1] CCTGTCCCTGTCGGCTCGGGTGCTCTG -5' <----------GGCTCGGGTGCTCTG [8-bp barcode3] TAGAGCATACGGCAGAAGACGAAC -5'
5'- CGCCGTCCAGCTGA [8-bp barcode2] CCGGGAGCTGCATGT ---------> 5'-CCGGGAGCTGCATGT [8-bp barcode1] TTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC [sgRNA-Spacer] CGGTGTTTC GTCCTTTCCACAA...CTAACAGCCTTGTATCGTATATGCA... -[WPRE]-[PuroR] -3' <---------GATTGTCGGAACATAGCATATACGT -5'C <- this is the extra C that won't anneal, not sure if this is a mistake
ATAC (ready to sequence): 5'- AATGATACGGCGACCACCGAGATCTACAC [8-bp barcode2] TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT GCCGTCCTCGATCGC [8-bp barcode1] GGACAGGGACAGCCGAGCCCACGAGAC [8-bp barcode3] ATCTCGTATGCCGTCTTCTGCTTG TTACTATGCCGCTGGTGGCTCTAGATGTG [8-bp barcode2] AGCAGCCGTCGCAGAGGTGCG TTGGTTGCGTGGCAGAGGCGGAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG [8-bp barcode1] CCTGTCCCTGTCGGCTCGGGTGCTCTG [8-bp barcode3] TAGAGCATACGGCAGAAGACGAAC -5' sgRNA (need another round of PCR, see the next step): 5'-CGCCGTCCAGCTGA [8-bp barcode2] CCGGGAGCTGCATGT [8-bp barcode1] TTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC [sgRNA-Spacer] CGGTGTTTC GTCCTTTCCACAA...CTAACAGCCTTGT G ATCGTATATGCA GCGGCAGGTCGACT [8-bp barcode2] GGCCCTCGACGTACA [8-bp barcode1] AAAAAGTTCAACTATTGCCTGATCGGAATAAAATTGAACGATAAAGATCGAGATTTTG [sgRNA-Spacer] GCCACAAAG CAGGAAAGGTGTT...GATTGTCGGAACA C TAGCATATACGT -5'
sgRNA: 5'- AATGATACGGCGACCACCGAGATCTACAC TCTTTCCCTACACGACGCTCTTCCGATCT [None/A/TA/GTA/NNNNNNNN]CGCCGTCCAGCTGA ----------------> 5'-CGCCGTCCAGCTGA [8-bp barcode2] CCGGGAGCTGCATGT [8-bp barcode1] TTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC [sgRNA-Spacer] CGGTGTTTC GTCCTTTCCACAA...CTAACAGCCTTGT G ATCGTATATGCA GCGGCAGGTCGACT [8-bp barcode2] GGCCCTCGACGTACA [8-bp barcode1] AAAAAGTTCAACTATTGCCTGATCGGAATAAAATTGAACGATAAAGATCGAGATTTTG [sgRNA-Spacer] GCCACAAAG CAGGAAAGGTGTT...GATTGTCGGAACA C TAGCATATACGT -5' <------------CACAAAG CAGGAAAGGTGTT [8-bp barcode3] TAGAGCATACGGCAGAAGACGAAC -5'
ATAC: 5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNN TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT GCCGTCCTCGATCGC NNNNNNNN GGACAGGGACAGCCGAGCCCACGAGAC NNNNNNNN ATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNN AGCAGCCGTCGCAGAGGTGCG TTGGTTGCGTGGCAGAGGCGGAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG NNNNNNNN CCTGTCCCTGTCGGCTCGGGTGCTCTG NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'Illumina P5 8-bp s5 ME gDNAME 8-bp s7 8-bp Illumina P7 barcode2 barcode1 barcode3 sgRNA: 5'-AATGATACGGCGACCACCGAGATCTACAC TCTTTCCCTACACGACGCTCTTCCGATCT ...CGCCGTCCAGCTGA NNNNNNNN CCGGGAGCTGCATGT NNNNNNNN TTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC [sgRNA-Spacer] CGGTGTTTC GTCCTTTCCACAA NNNNNNNN ATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG AGAAAGGGATGTGCTGCGAGAAGGCTAGA ...GCGGCAGGTCGACT NNNNNNNN GGCCCTCGACGTACA NNNNNNNN AAAAAGTTCAACTATTGCCTGATCGGAATAAAATTGAACGATAAAGATCGAGATTTTG [sgRNA-Spacer] GCCACAAAG CAGGAAAGGTGTT NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'Illumina P5 TruSeq Read 1 |8-bp 8-bp CROP-seq sgRNA scaffold 8-bp Illumina P7 variable bases:barcode2 barcode1 barcode3 None, A, TA, GTA or 8 Ns
5'- CACCGTCTCCGCCTC AGATGTGTATAAGAGACAG -------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNN AGCAGCCGTCGCAGAGGTGCG TTGGTTGCGTGGCAGAGGCGGAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG NNNNNNNN CCTGTCCCTGTCGGCTCGGGTGCTCTG NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'
5'- CTGTCTCTTATACACATCT GCCGTCCTCGATCGC ------------------------------------------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNN AGCAGCCGTCGCAGAGGTGCG TTGGTTGCGTGGCAGAGGCGGAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG NNNNNNNN CCTGTCCCTGTCGGCTCGGGTGCTCTG NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC -------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG NNNNNNNN AGCAGCCGTCGCAGAGGTGCG TTGGTTGCGTGGCAGAGGCGGAG TCTACACATATTCTCTGTC XXX...XXXGACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG NNNNNNNN CCTGTCCCTGTCGGCTCGGGTGCTCTG NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'
5'- AATGATACGGCGACCACCGAGATCTACAC NNNNNNNN TCGTCGGCAGCGTCTCCACGC AACCAACGCACCGTCTCCGCCTC AGATGTGTATAAGAGACAG XXX...XXXCTGTCTCTTATACACATCT GCCGTCCTCGATCGC NNNNNNNN GGACAGGGACAGCCGAGCCCACGAGAC NNNNNNNN ATCTCGTATGCCGTCTTCTGCTTG -3' <------GACAGAGAATATGTGTAGA CGGCAGGAGCTAGCG -5'
5'- ACAC TCTTTCCCTACACGACGCTCTTCCGATCT -----------------------------------------------------------------------------------------------------------------------------------------------------> 3'-TTACTATGCCGCTGGTGGCTCTAGATGTG AGAAAGGGATGTGCTGCGAGAAGGCTAGA ...GCGGCAGGTCGACT NNNNNNNN GGCCCTCGACGTACA NNNNNNNN AAAAAGTTCAACTATTGCCTGATCGGAATAAAATTGAACGATAAAGATCGAGATTTTG [sgRNA-Spacer] GCCACAAAG CAGGAAAGGTGTT NNNNNNNN TAGAGCATACGGCAGAAGACGAAC -5'